For decades, finding clues to substance addiction has been much like searching for a needle in a haystack. But researchers may finally be honing in on specific genes tied to all types of addictions – and finding that some of the same genes associated with alcohol dependence are also closely linked with addictions to nicotine, cocaine, opoids, heroin and other substances.
In a new landmark paper to be published in the April 2009 issue of Nature Genetics Reviews , addiction experts at the University of Virginia Health System and the University of Michigan present new insights into the significant progress made within the last several years in understanding the genetics of addiction.
“Addiction researchers have found that several genes are linked with multiple addictions,” says co-author Ming Li, Ph.D., professor of psychiatry and neurobehavioral sciences at the UVA School of Medicine. “So, we’re narrowing the scope to specific genetic targets. Once researchers can pinpoint exact genetic variants and molecular mechanisms, then we can create much more effective, even personalized, treatments for individuals addicted to a variety of substances.”
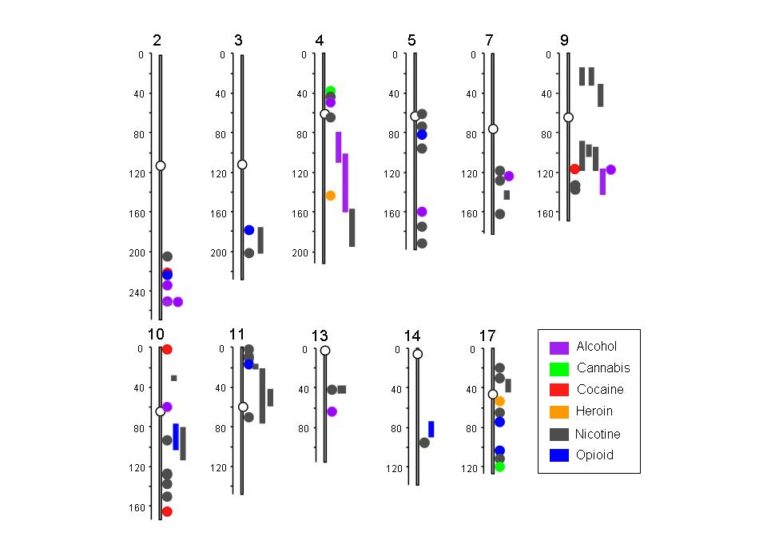
In the paper, which in part serves as a comprehensive guide to the existing body of addiction genetics research, Li and co-author Margit Burmeister, Ph.D., professor of psychiatry and human genetics at the University of Michigan, present a summary of specific genomic locations on 11 chromosomes where addictions to alcohol, cannabis, cocaine, heroin, nicotine and opoids are clustered together.
“The comparison of peaks for addictions to multiple substances on certain chromosomal locations confirms that genetic vulnerability to different substances overlaps, in part,” Li says. He further points out that variants in several genes, including aldehyde dehydrogenases, GABRA2 , ANKK1 , and neurexins 1 and 3, have already been associated with addictions to multiple drugs.
In recommending a future direction for research into the genetics of addiction, Li suggests focusing on CHRNA5, CHRNA3 and CHRNB4 clusters, among other variants. “The exact nature of the gene variants and how they function are still unknown, so functional studies as well as studies using additional ethnic population samples may be quite revealing.”