CHARLOTTESVILLE, Va. – University of Virginia Health System researchers are the first in the world to develop a new and faster method to track major infection-causing “superbugs” – a major key in preventing the spread of deadly infections.
Their research, published in the November/December 2011 issue of the online journal mBio , comes at a critical time.
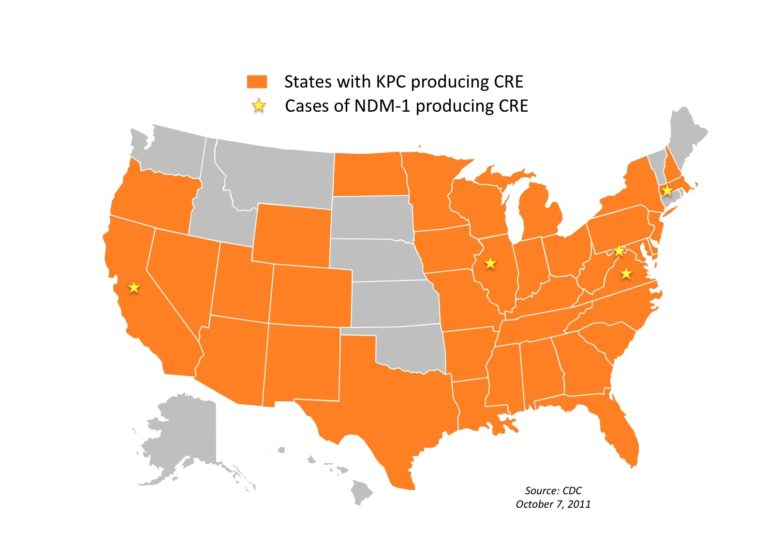
Several newly discovered genes such as KPC ( Klebsiella pneumoniae carbapenemase) and NDM (New Delhi metallo-betalactamase) have experts on alert for several reasons:
their ability to easily transform bacteria into antibiotic-resistant superbugs their unprecedented ability to cross over into different strains and species of bacteria the ease with which they can transfer to highly infectious bacteria, such as Salmonella and cholera, and the potential for these genes to easily establish themselves undetected in our environment.
“When you’re in a race against time to halt the spread of these life-threatening infections, the traditional methods of detection and tracking are very difficult and frankly take too long,” says UVA Medical Center Epidemiologist Costi Sifri, MD , study principal investigator.
Decoding deadly hospital-acquired infections
The focus of their study was KPC, a gene Sifri calls “perhaps the most important new resistance gene of the millennium.” KPC has the ability to easily hitch a ride on movable genetic elements called plasmids that then spread between pathogens, making them highly infectious and deadly.
Through their new method, Sifri’s research team was able to quickly track the movement of plasmids carrying KPC between bacteria and understand the spread of KPC bacteria at the molecular level.
While traditional methods take several weeks or months to do this analysis and can only be performed in specialized research laboratories, the method developed by the UVA team takes only one to two days and can be performed by most modern microbiology laboratories.
“We developed and successfully used a novel technique to rapidly track the gene’s movement between patients and bacteria,” says lead author Amy Mathers, MD, assistant professor in the UVA School of Medicine’s Division of Infectious Diseases and International Health .
“We were able to show that all cases in a cluster of hospital infections could be traced back to a single patient. Most cases were linked by movement of the KPC gene between different bacteria.”
Armed with this information, the research team hopes to develop new strategies and tools to prevent the spread of these highly resistant bacteria to other patients.
The pathogens associated with this new gene are called CRE, or Carbapenem- Resistant Enterobacteriaceae, and include E. coli and Klebsiella , among others. Infections caused by CRE are associated with very high mortality rates, Sifri says.
A global health threat
“These CRE pathogens are widely considered a major healthcare threat in the United States and worldwide,” says Sifri. “Monitoring their origin and spread is critical to preventing serious outbreaks in hospitals, other healthcare facilities, and potentially the general population.”
In fact, another drug-resistant gene called NDM emerged last year as a cause of significant disease in India and may be acquired through drinking water. The pathogen spread quickly and was carried to the U.S. and Great Britain.
“A class of antibiotics called carbapenems has become essential for the treatment of many infections in hospitals today, and their potential loss as a reliably effective class of antibiotics is a serious public health threat,” says Sifri.
“The more rapidly we can detect CRE and understand movement of the KPC gene in hospitalized patients, the more quickly we can intervene to prevent CRE spread to other patients and other strains of bacteria. This is fundamentally important for maintaining the effectiveness of this important class of antibiotics.”
The study “Molecular Dissection of an Outbreak of Carbapenem-Resistant Enterobacteriaceae Reveals Intergenus KPC Carbapenemase Transmission through a Promiscuous Plasmid,” can be found online in the mBio November/December 2011 issue.